Services/Equipment
Preparation of 16S (and all other) amplicons for Illumina sequencing
We offer Fluidigm preparation of custom amplicons!
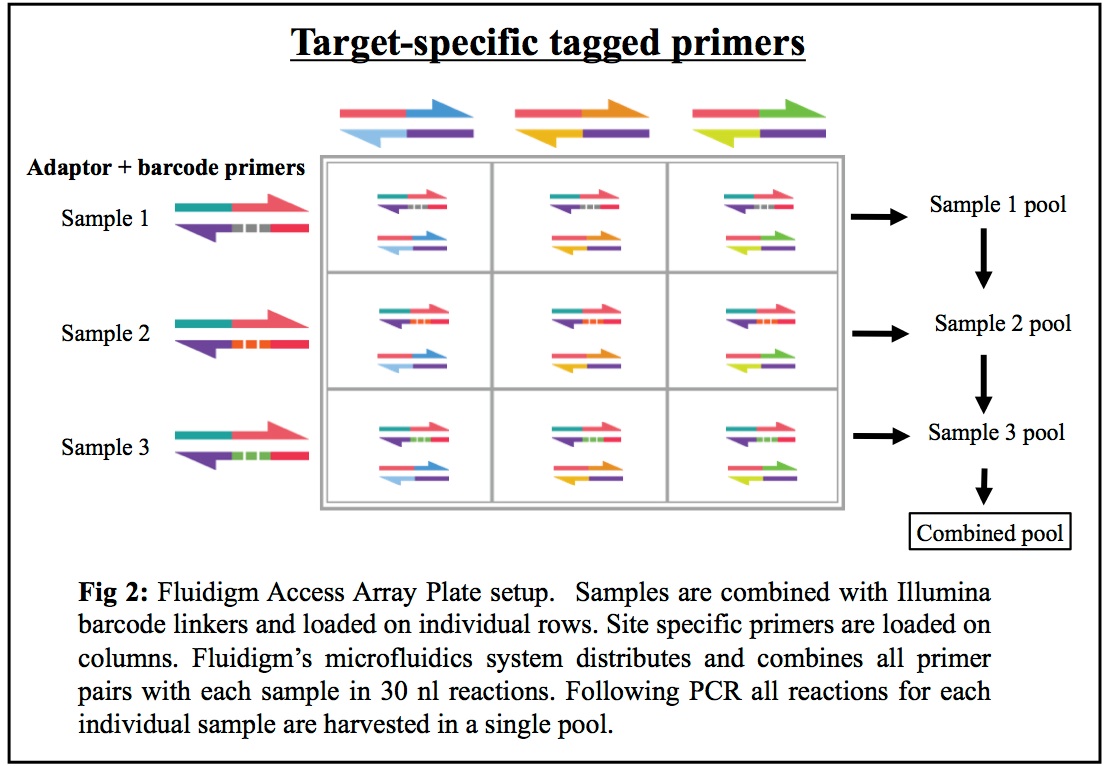
Our combined Fluidigm+MiSeq protocol is a cost-effective system allowing researchers to sequence up to 1500 different metagenomic 16S rRNA gene samples (or any other amplicon) against up to 24 different genes/targets, using a streamlined and repeatable microfluidics process that minimizes PCR errors and cross-contamination while maximizing your potential understanding of the mixed communities being studied.
How the Fluidigm Platform Works:
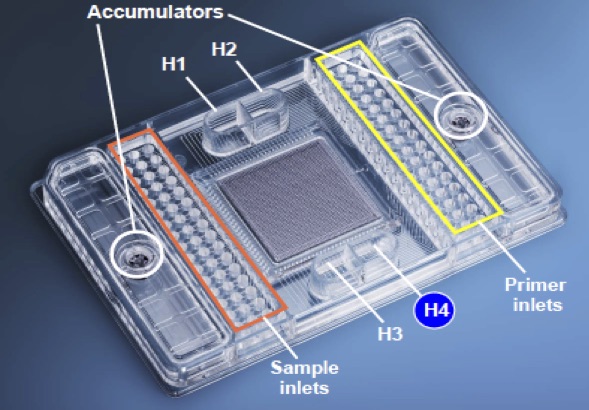
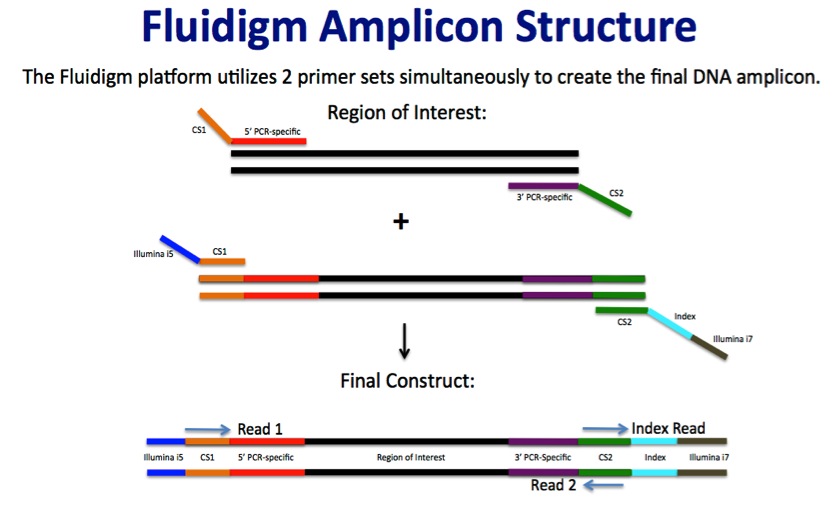
Fluidigm Amplicon Design

Common Primers Offered for Amplicon Sequencing
We offer hundreds of different PCR primers for 16S rRNA as well as 18S, eukaryotic, ITS, archaeal, and other functional gene targets. Our list is continuously being upgraded, so please check with us for the latest primer selections.
|
Target |
Primer Name |
Primer Sequence (5' to 3') |
Expected FINAL* Product Length (nt) |
|
16S V1-V3 |
V1-V3 F28 |
GAGTTTGATCNTGGCTCAG |
643 |
|
|
V1-V3 R519 |
GTNTTACNGCGGCKGCTG |
|
|
16S V3-V5 |
V3-V5 F357 |
CCTACGGGAGGCAGCAG |
694 |
|
|
V3-V5 R926 |
CCGTCAATTCMTTTRAGT |
|
|
16S V4 |
V4 515F |
GTGCCAGCMGCCGCGGTAA |
252 |
|
|
V4 806R |
GGACTACHVGGGTWTCTAAT |
|
| 16S V4 (new) | V4 515F (new) | GTGYCAGCMGCCGCGGTAA | 252 |
| V4 806R (new) | GGACTACNVGGGTWTCTAAT | ||
|
Archaea |
Arch349F |
GYGCASCAGKCGMGAAW |
528 |
|
|
Arch806R |
GGACTACVSGGGTATCTAAT |
|
|
Eukaryotic 18S |
F566Euk |
CAGCAGCCGCGGTAATTCC |
765+ |
|
|
R1200Euk |
CCCGTGTTGAGTCAAATTAAGC |
|
|
Eukaryotic 18S |
Euk_1391F |
GTACACACCGCCCGTC |
200-280 |
|
|
EukBr-7R |
TGATCCTTCTGCAGGTTCACCTAC |
|
|
ITS1-ITS4 |
ITS1 |
TCCGTAGGTGAACCTGCGG |
580+ |
|
|
ITS4R |
TCCTCCGCTTATTGATATGC |
|
|
ITS3-ITS4 |
ITS3F |
GCATCGATGAAGAACGCAGC |
462+ |
|
|
ITS4R |
TCCTCCGCTTATTGATATGC |
|
How to Submit Samples for amplification and sequencing on the Fluidigm+MiSeq/HiSeq:
The success of the amplicon construction and sequencing depends heavily on the integrity and purity of the DNA. Degraded or fragmented DNA produces weak amplicons with amplification artifacts. Even if just one of the DNA samples is fragmented, the artifacts produced by amplification can affect the results of the entire pool. DNA with humic acids or other contaminants that interfere with amplification will also produce poor results. Removal of RNA during DNA purification is preferred.
Please, check as many of your DNA samples as possible on a 1% gel and evaluate the integrity of the DNA:
Run an aliquot (~50 to 100ng) of the DNA on a 1% agarose gel next to a DNA ladder. This picture can be used to evaluate integrity of the DNA as well as presence/absence of RNA.
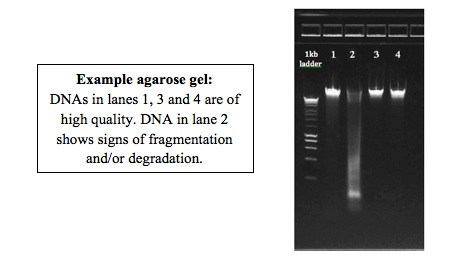
For Fluidigm submission of metagenomic/16S assays we require at least 2 ng/ul quantitated by Qubit or Picogreen. Submit at least 10 ul of each DNA sample. Please contact our sister unit, Functional Genomics, for the latest primer information, submission guidelines and submission form.
Functional Genomics Unit
Mark Band, Ph.D - Director
356 Edward R. Madigan Laboratory
1201 W. Gregory Drive
Urbana, IL 61801
Phone: (217) 244-3930 FAX: (217) 265-5066
Email: markband@illinois.edu
The DNA Services lab is available to discuss all sequencing options and pricing (contact information below).
MiSeq Pricing Information »
Please contact Dr. Alvaro Hernandez, Director of DNA Services ( aghernan@illinois.edu) at 217-244-3480 or Chris Wright, Assistant Director of DNA Services (clwright@illinois.edu) at 217-333-4372 to discuss ways the staff can be of assistance in achieving your project goals or to receive a quote for your project.
All work performed by the Roy J. Carver Biotechnology Center (CBC) should be acknowledged in scholarly publications, posters, and presentations. Proper recognition allows us to measure the impact of our work and supports our initiatives in obtaining sponsored funding. In addition, any CBC personnel who make a substantial intellectual or experimental contribution are deserving of further recognition as co-author.