Services/Equipment
Illumina iScan

The Functional Genomics Laboratory is equipped with an Illumina iScan for SNP genotyping and Human Methylation arrays. Illumina arrays are based on oligo coated beads each unique to a specific SNP locus. The beads are randomly distributed and located in 3 micron wells etched in a slide.
Beadarrays include all commercially available Illumina Arrays as well as Agricultural Consortium Arrays. https://www.illumina.com/areas-of-interest/agrigenomics/consortia.html
For a full list of available products or an overview of the beadarray technology see the Illumina website at http://www.illumina.com/. Custom SNP design is also available through Illumina.


Hybridized high density Infinium slides are scanned in the Illumina iScan
Image of a single bead cluster showing genotypes for one DNA sample.
SNP genotypes: AA (green), BB (red), AB (yellow)
Analysis of bead array signals is carried out using the Illumina GenomeStudio software.
Genotype calls are exported in tabular format compatible for linkage or association studies with third party programs.
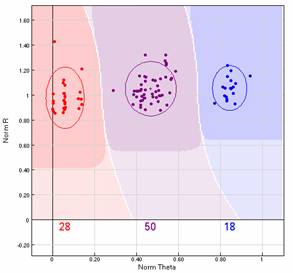
SNP cluster showing genotype calls for 96 DNA samples.
For additional information and specific project pricing contact Dr. Mark Band (markband@illinois.edu)217-244-3930.
All work performed by the Roy J. Carver Biotechnology Center (CBC) should be acknowledged in scholarly publications, posters, and presentations. Proper recognition allows us to measure the impact of our work and supports our initiatives in obtaining sponsored funding. In addition, any CBC personnel who make a substantial intellectual or experimental contribution are deserving of further recognition as co-author.
