Services/Equipment
Services
- Protein identification by high resolution LC-MS/MS
- Intact mass analysis
- Quantitative global and targeted proteomics
- Post-translational modification analysis
- Intact Protein Characterization (both native and denatured states)
- Sample preparation from gel bands, beads, cells, tissue, biofluids, etc.--Now taking Biosafety Level II samples with prior agreement!
Equipment
Main equipment includes 3 mass spectrometers: 1) Thermo Q Exactive HF-X Quadrupole-Orbitrap, 2) Thermo Fusion Tribrid Orbitrap, and 3) Thermo Q Exactive Ultra High Mass Range (UMHR) that are featured below.
Thermo Fusion Orbitrap Tribrid Mass
Spectrometer with ETD Fragmentation 
The versatile Fusion Orbitrap Tribrid mass spectrometer accommodates a variety of analysis types. This instrument is used for everything from routine protein identification to post-translational modification identification and localization using one of its three fragmentation mechanisms (CID, HCD,and ETD). Its tribrid design also permits higher order tandem MS scans, meaning that we can make use of newer isobaric reagents such as iTRAQ and TMT for label-based protein quantitation in addition to more traditional methods such as SILAC. The facility also has a Advion TriVersa NanoMate robot to facilitate intact protein analyses using the Fusion.
Thermo Q Exactive HF-X Mass Spectrometer 
with BioPharma Option
The QE HF-X mass spectrometer with BioPharma option can be used for high resolution accurate mass (HRAM) measurements in both bottom-up and top-down proteomic analyses. In conjunction with chromatographic separation
using an Ultimate 3000 nLC system, the highly sensitive HF-X can detect thousands of proteins from digested cell lysate in just one hour. The high resolution (up to 240k) and extended mass range of this instrument (up to 8,000 m/z) also make it useful for characterization of mid-sized proteins. An important application utilizing the HF-X is identification of proteins such as antibodies with modifications including oxidation and complex glycosylation. The HF-X is also useful for quantitation workflows including parallel reaction monitoring (PRM) and label-free analyses.
Thermo Q Exactive UHMR
Mass Spectrometer 
The Q Exactive Ultra High Mass Range (UHMR) Mass Spectrometer
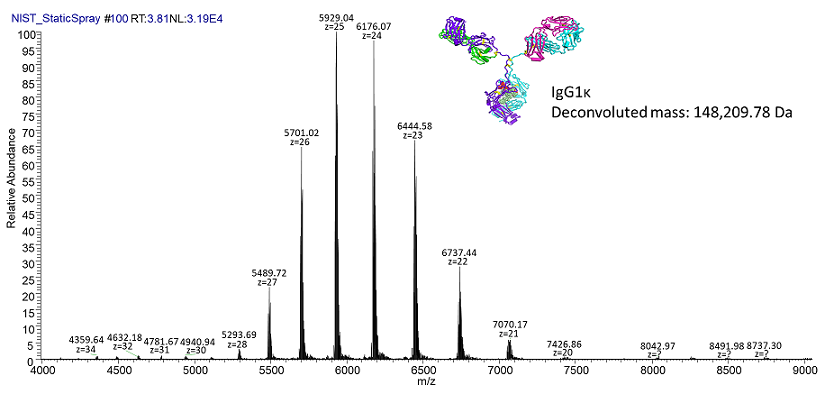
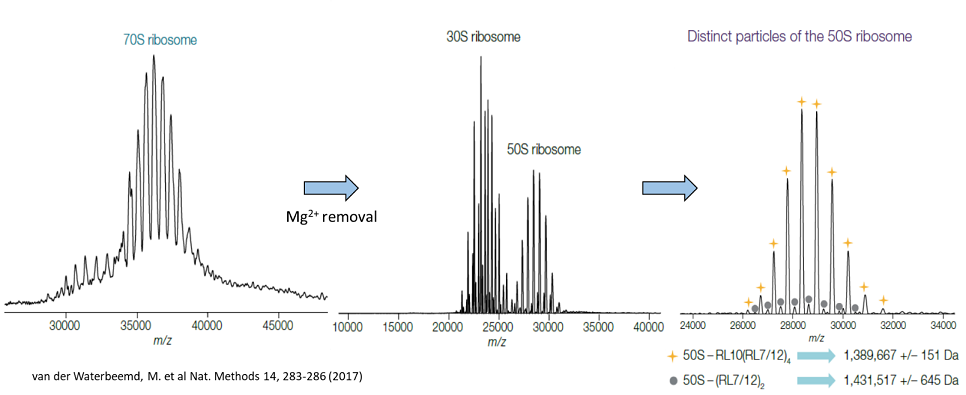
is the newest addition to the facility. With a mass range of 350 to 80,000 m/z, this instrument is a powerful tool for studying proteins and protein complexes up to several megadaltons. Importantly, proteins and protein complexes can be analyzed in their native states, permitting characterization of non-covalent protein-protein interactions and protein-ligand binding. In-source trapping and HCD fragmentation capabilities also permit top-down analyses in which protein complexes are broken apart to examine individual subunits. In addition to a high mass range, the UHMR also boasts high resolution (up to 200k at 400 m/z) that can be used to differentiate protein complexes with similar compositions from one another.
Analyses using the UHMR are supported by an Advion TriVersa NanoMate robot, which allows robust ionization of small sample amounts in the uL range. The Thermo BioPharma Finder software suite is also available for deconvolution and sequencing of native top-down experiments. With training by Proteomics Core staff, university affiliated researchers can use the UHMR mass spectrometer and Advion NanoMate.
All work performed by the Roy J. Carver Biotechnology Center (CBC) must be acknowledged in scholarly publications, posters, and presentations. Proper recognition allows us to measure the impact of our work and supports our initiatives in obtaining sponsored funding. In addition, any CBC personnel who make a substantial intellectual or experimental contribution are deserving of further recognition as co-author.
