10x Single Cell Submission Requirements and Other Information:
Our facility offers a full range of services for library construction and sequencing with the NovaSeq 6000, MiSeq, Oxford Nanopore GridION and the 10x Genomics Chromium. The information below is for 10x Genomics Single Cell work only. Please return to our main Sample Submission page for all other sample types.
There is a suite of different library prep options from 10x Genomics:
-
sc/snRNASeq: polyA capture from 3' and 5' based kits.
-
Add-ons to the RNASeq libraries include V(D)J, Cell Surface Protein, and CRISPR Screening
-
-
snATACSeq: Epigenetic profiling of open chromatin
-
Multiome: snRNA + snATAC on the same nuclei
-
Fixed RNA: Single cell/nuclei RNAseq for fixed human or mouse samples only. This kit is probe based and can offer cost-savings on larger projects.
Contact Information:
-
All inquiries about 10x Single Cell should be directed to ngsequencing@illinois.edu. This email will copy both Chris Wright and Alvaro Hernandez so you receive the quickest reply possible.
-
Timing is critical. If you are considering single cell work, the first thing you should do is contact the lab to reserve dates. At a minimum, please contact the DNA Services lab BEFORE you begin growing your cells or your experimental study animals to schedule test counts and setup dates.
- Scheduling usually occurs:
-
10x single cell test counts: ~1-2+ weeks from when you contact us
-
10x library construction: ~4-8 weeks from when you contact us
-
-
Below, you'll find a host of details and links that you can refer to throughout this process.
Basic Information:
-
10x Genomics Single Cell libraries can provide incredibly powerful data by allowing interrogation of highly heterogeneous tissues on an individual cell level. Click the 10x Genomics link to read more about the technology and it's various applications.
-
The general process is as follows:
-
Determine experimental needs --> meet with us, and with HPCBio if you need analysis help.
-
Reserve dates for both test counts and full experiment.
-
Test tissue dissociation protocols.
-
Perform test counts with DNA Services to see how your cells or nuclei look (viability, debris, etc.)
-
Adjust dissociation method and repeat test counts as needed.
-
Perform full experiment--extract cells/nuclei and bring to lab on previous scheduled dates. Libraries are constructed.
-
After library construction, sequencing is typically started within ~ 1 week.
-
**Please email to schedule test counts with us as soon as possible, at least 2 weeks in advance, as single cell work is VERY popular and employees are already scheduled with current in-queue projects.
Cell Dissociation:
-
We receive cells from nearly every source imaginable. While there may not be an exact protocol for cell dissociation of your specific tissue type, there are hundreds of protocols available on the 10x website, Worthington website, and Miltenyi website that can be adapted.
-
The Biotechnology Center CMtO Core facility offers the GentleMACS Dissociator instrument for use by University of Illinois faculty, students, and staff. You are responsible for purchasing the appropriate Miltenyi dissociation kit and tubes for your use. There is currently no charge to use the GentleMACS Dissociator, you would check it out from Flow for use in your lab.
-
Miltenyi makes a Tissue Storage Solution that provides for optimized storage of fresh organ and tissue samples without showing background effects like cell activation or apoptosis induction.
Nuclei Isolation:
-
We have had excellent success across a variety of samples (fish, fox, mouse) with the Nuclei Isolation Kit from 10x Genomics. This kit is compatible with snRNA, snATAC, snMultiome, and Fixed RNA kits.
-
snRNASeq is the only 10x application that can start with either nuclei or cells. See comparison data between cells and nuclei here.
-
Using nuclei instead of cells has several advantages:
-
Reducing bias introduced by various cell dissociation methods.
-
Minimizing trascriptional changes during dissociation/stress.
-
Capturing newly synthesized mRNA in temporal studies.
-
Allows for freezing of tissue samples at collection and batch isolation of processing of all samples at one (also reducing bias).
-
One drawback to using nuclei is that these libraries tend to have slightly higher levels of background RNA, ie: not "in-cell" RNA, which is removed bioinformatically.
-
Cell Requirements, Tips, and Tricks:
-
Do I need replicates?: In general, yes. When 10x Genomics first started, people generated and published with one library per treatment. Now, reviewers are asking for replicates (3). Our current recommendation is that you don't need replicates if you are generating preliminary data for a grant proposal. However, if you are trying to publish the results, you should plan for 3 biological replicates.
-
Starting cell concentration should be at least 1,000 cells/ul (1M cells/ml) or more. We request 1M cells total, transported to us in a 1.5ml centrifuge tube. Only bring cells in a 1.5 or 2.0ml centrifuge tube. We will count and concentrate cells to the appropriate concentration for the protocol you are using. There are modified protocols for limited cells, please ask us if this is your case, we have worked with cells that could not be counted, we can make your project work too!
-
Please see this Cell Prep Guide for information on compatible buffers. Final media should not contain excessive amounts of EDTA (> 0.1mM), calcium or magnesium (> 3mM) as those components will inhibit the reverse transcription reaction. Any surfactants (Tween-20, etc) should also be avoided as they may interfere with GEM generation. Different applications require different final buffers--check with us!
-
Cells do not need to be flow sorted prior to 10x single cell library construction. You should sort only if you need to target or remove a particular cell population, as sorting takes time, dilutes, and stresses the cells. If you will be flow sorting first, you will need to copy both CMtO staff and DNA Services staff to set up the project dates and times. Do not sort nuclei; you must sort cells then perform nuclei isolation.
-
Elimination of dead cells is critical. Cell viability should be >70%, but >80% is better, and over 90% is ideal. This is true even if isolating nuclei. Your starting cell suspension will be counted, washed, and recounted upon arrival with live/dead measure included. If viability is below 70%, we will recommend that you repeat cell collection as background RNA levels will be high.
-
If cell suspensions cannot be obtained with >70% viability, we recommend you first try using a Flowmi 40 μm cell strainer. These filters are incredibly handy and easy to use, just put them on the tip of your 1000μl pipette tip and slowly push your suspension through to a 1.5ml tube. These will often clean up your sample nicely.
-
If cell suspensions still cannot be obtained with >70% viability, you'll need to use the Miltenyi Biotec Dead Cell Removal kit with MS columns (catalog # 130-090-101 / 130-042-201), which removes dead and dying cells. You must purchase both the kits and the columns, linked in the previous sentence. We can also do this for a fee.
-
The magnet and single-tube stand needed to use the Miltenyi Dead Cell Removal kit with MS columns can be checked out from the Biotechnology Center CMtO Core facility.
-
We also have an octo-magnet and stand in our DNA Services BL2 10x facility, 221 ERML, that you may use when you bring cells for counting. In this case, we recommend you add the Dead Cell binding beads in your lab and use your transport time to our facility as the incubation time. Note that incubation of cells and binding beads is at room temperature, do not put your cells on ice for this step.
-
-
You may also need to perform an RBC lysis step on your cells. This is the 10x answer for why and how. If RBC lysis is needed, we recommend purchasing the 10x Genomics recommended RBC lysis buffer. Researchers who have tried homemade lysis buffer have significantly delayed their projects.
The numbers:
-
Below you'll find some general information, but please do meet with us to discuss your project rather than trying to figure it all out by yourself.
-
A library can be made from 500-20,000 cells, depending upon your project needs. HT RNA libraries, if you have 96+ samples, can capture up to 20k cells per library. Cell populations with more heterogeneity will typically require a higher total cell capture. Most libraries we construct now target 8-10k cells per sample.
-
Doublets increase linearly with total cells, from ~0.2% at 500 cells, to ~8% at 20,000 cells with the latest GEM-X kits.
-
We recommend capturing at least 3,000 cells or more. Libraries capturing less than 3,000 cells have much greater variability in actual cells captured (+/- 50% or more) and lower reproducibility. Variability of actual capture at or above 3,000 cells should be ~30% or less (ie: 2,100-3,900 for a 3,000-cell target). Keep in mind, it is significantly easier to resequence a library for more depth than to repeat an experiment for more cells. If you cut costs, cut cost with sequencing, capture as many cells as you can afford.
-
Sequencing recommendation varies by cell and species type. PBMCs require much less sequencing, ie: 25-50k reads, compared to highly transcriptional active cells which could need over 100k reads before the majority of genes are detected.
-
(C) 10x Genomics: Sequencing Saturation examples:
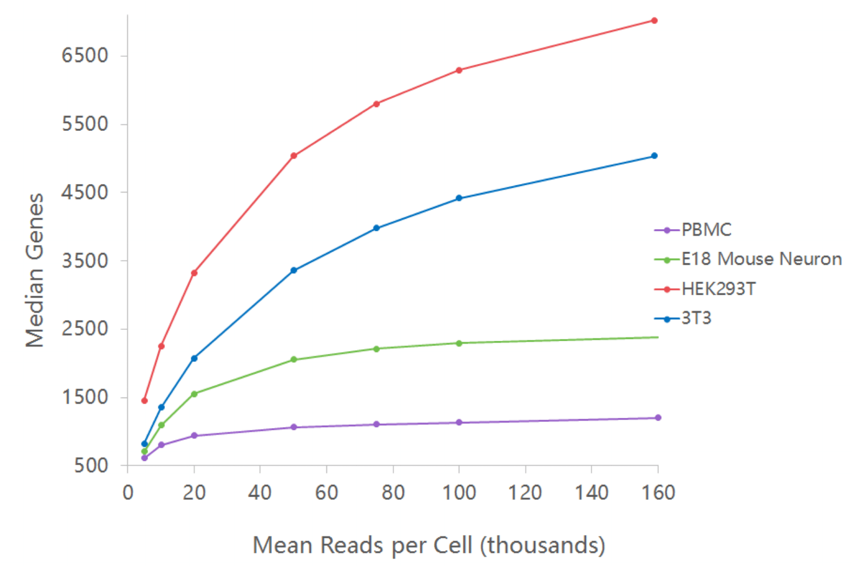
-
While all protocols for library preparation will be followed, due to differences in sample types submitted for 10x single cell, no guarantee is given that your cell type will behave according to typical performance.
-
Sequencing:
-
For 10x 3' transcriptome V3.1 GEM-X single cell kits, we generally recommend at least 50k sequencing reads per targeted cell. See image above.
Example costs for various projects
A full listing of costs can be found on our pricing page. Below are example quotes for a pilot study, and 16 sample project with either polyA capture or Fixed RNA approach.
2 sample pilot study, scRNA (polyA capture) GEM-X library. Used for preliminary data for grant proposals only; publications require 3x replicates:
| Service | Price/Item | Qty | Cost |
| 10X Single Cell RNA Library | $2,750.00 | 2 | $5,500.00 |
| Nova X 10B paired-reads 300-cycle, per lane | $2,190.00 | 1 | $2,190.00 |
| Subtotal: | $7,690.00 |
16 sample project, snRNA (polyA capture), GEM-X:
| Service | Price/Item | Qty | Cost |
| 10X Single Cell RNA Library (8+ at once) | $2,050.00 | 16 | $32,800.00 |
| 10x Nuclei Isolation Kit | $2,000.00 | 2 | $4,000.00 |
| MiSeq Titration | $397.00 | 1 | $397.00 |
| Nova X 25B paired-reads 300-cycle, per lane | $3,290.00 | 3 | $9,870.00 |
| Subtotal: | $47,067.00 |
16 sample project, Fixed RNA (human or mouse only, probe based):
| Service | Price/Item | Quantity | Cost |
| 10x Fixed RNA Sample Fixation, Set up fee | $448.00 | 2 | $896.00 |
| 10x Fixed RNA Sample Fixation, each | $99.00 | 16 | $1,584.00 |
| 10x Fixed RNA, 1 channel (up to 16 lib) | $14,210.00 | 1 | $14,210.00 |
| 10x Nuclei Isolation Kit* | $2,000.00 | 2 | $4,000.00 |
| Nova X 25B paired-reads 300-cycle, per lane | $3,290.00 | 1 | $3,290.00 |
| Subtotal: | $23,980.00 |
*Minimum of 2-5 Nuclei isolation kits are needed for 16 samples, based on tissue source and size:
(C) 10x Genomics: Expected yield across different tissue types with 10x Genomics Nuclei Extraction Kit:
Scheduling Meetings, Test Counts, and Experiments
Scheduling Test Counts:
-
There is no charge to run test counts, either independently or with assistance from DNA Services employees. None. All consumables for counting are also included. Please, email us to schedule these test counts as soon as you know you are doing a single cell project, at least 2-4 weeks in advance. 10x Genomics libraries are very popular and we want to be available when you are ready!
-
Any new cell type should have at least one successful test count prior to the full experiment (library construction). Most projects require just 1-2 test counts before they have achieved the viability and quality required for 10x single cell experiments.
-
We use the Nexcelom (now Revvity) K2 automated cell counter for single cell libraries. This instrument counts using both brightfield and fluorescent imaging with live/dead count using AO/PI. It is very easy to use, flexible, and consistent. Settings can be changed for each sample type you have and saved as your profile for easy repeat counting.
Recent Single Cell Publications from on campus:
•Single cell RNA-sequencing reveals the complete temporal sequence of transcription factors that pattern Drosophila medulla neuroblasts. Hailun Zhu, Sihai Dave Zhao*, Alokananda Ray, Yu Zhang and Xin Li*. In preparation.
•Single-cell analyses of the corneal epithelium: Unique cell types and gene expression profiles. Surabhi Sonam, Sushant Bangru, Kimberly J. Perry, Auinash Kalsotra, Jonathan J. Henry. bioRxiv 2020.08.06.240036.
–https://doi.org/10.1101/2020.08.06.240036
•Meta-analysis of honey bee neurogenomic response links Deformed wing virus type A to precocious behavioral maturation. Traniello, I.M., Bukhari, S.A., Kevill, J. et al. Sci Rep 10, 3101 (2020).
–https://doi.org/10.1038/s41598-020-59808-4
•Cellular plasticity balances the metabolic and proliferation dynamics of a regenerating liver. Ullas V. Chembazhi, Sushant Bangru, Mikel Hernaez, Auinash Kalsotra
–https://doi.org/10.1101/2020.05.29.124263
•A common pattern of influenza A virus single cell gene expression heterogeneity governs the innate antiviral response to infection. J. Cristobal Vera, Jiayi Sun, Yen Ting Lin, Jenny Drnevic, Ruian Ke, Christopher B. Brooke. bioRxiv 858373.
–https://doi.org/10.1101/858373
•Progesterone Receptor Serves the Ovary as a Trigger of Ovulation and a Terminator of Inflammation. Park CJ, Lin PC, Zhou S, et al. Cell Rep. 2020;31(2):107496.
–https://doi.org/10.1016/j.celrep.2020.03.060
Please contact Dr. Alvaro Hernandez, Director of DNA Services (aghernan@illinois.edu) at 217-244-3480 or Chris Wright, Associate Director of DNA Services (clwright@illinois.edu) at 217-333-4372 to discuss ways the staff can be of assistance in achieving your project goals or to receive a quote for your project, submission forms, grant support or other information needed.
All work performed by the Roy J. Carver Biotechnology Center (CBC) should be acknowledged in scholarly publications, posters, and presentations. Proper recognition allows us to measure the impact of our work and supports our initiatives in obtaining sponsored funding. In addition, any CBC personnel who make a substantial intellectual or experimental contribution are deserving of further recognition as co-author.