News
February 2024
The PacBio REVIO is here!
This cutting-edge long-read sequencing instrument offers exciting new capabilities. Its applications span a wide range, including de novo genome assembly for both eukaryotic and prokaryotic species, the most cost-effective genome resequencing using long reads, sequencing of long amplicons including full-length 16S, and full-length mRNA sequencing from bulk or single-cell RNAseq samples.
The standout advantage of the REVIO lies in its significantly reduced sequencing cost. Compared to the Sequel, it produces three times the data for the same cost, making it an best choice for researchers for their long-reads needs.
We eagerly anticipate discussing how long-read sequencing can enhance your research endeavors. Feel free to reach out for further details! 😊

September 2023
The Single-Cell and Spatial Transcriptomics workshop was a huge success. This workshop included presentations of all the latest applications, technologies, and instrumentation.
A recording of all the lectures can be accessed in the link below. If you are considering utilizing single-cell or spatial transcriptomics, please watch the recording to become familiarized with the most up to date information.
https://mediaspace.illinois.edu/media/t/1_n3a5pevn
July 2023
The NovaSeq X Plus is here!
The NovaSeq X Plus is the latest and most advanced DNA sequencing instrument currently available, able to sequence up to 20 billion DNA fragments in just one day. To translate this to samples, the output is 6 Tb of genomic information, equivalent to 2,000 human genomes, 6,000 soybean genomes, or 1 million bacterial genomes!
In addition, The X Plus system reduces current sequencing costs by up to **50%**, further enabling PIs to extend their funding into new or more complex research, utilizing genomics, transcriptomics, epigenomics, single-cell multiomics, spatial transcriptomics and proteomics, and metagenomics at a significantly lower cost compared to any other sequencing technology.

April 29 2022
Watch our latest seminar, with updates on all our capabilities and equipment, including details about single-cell and spatial transcriptome, in this link (it may take 2-3 minutes to start)=
https://uofi.box.com/s/9xcgnjlkisrdoprsnslc04c8fhj4jz12
10x Single Cell is booming! Below is just a selection of publications from our on-campus researchers. Contact us to get started (Oct 2020)
•Single cell RNA-sequencing reveals the complete temporal sequence of transcription factors that pattern Drosophila medulla neuroblasts. Hailun Zhu, Sihai Dave Zhao*, Alokananda Ray, Yu Zhang and Xin Li*. In preparation.
•Single-cell analyses of the corneal epithelium: Unique cell types and gene expression profiles. Surabhi Sonam, Sushant Bangru, Kimberly J. Perry, Auinash Kalsotra, Jonathan J. Henry. bioRxiv 2020.08.06.240036.
–https://doi.org/10.1101/2020.08.06.240036
•Meta-analysis of honey bee neurogenomic response links Deformed wing virus type A to precocious behavioral maturation. Traniello, I.M., Bukhari, S.A., Kevill, J. et al. Sci Rep 10, 3101 (2020).
–https://doi.org/10.1038/s41598-020-59808-4
•Cellular plasticity balances the metabolic and proliferation dynamics of a regenerating liver. Ullas V. Chembazhi, Sushant Bangru, Mikel Hernaez, Auinash Kalsotra
–https://doi.org/10.1101/2020.05.29.124263
•A common pattern of influenza A virus single cell gene expression heterogeneity governs the innate antiviral response to infection. J. Cristobal Vera, Jiayi Sun, Yen Ting Lin, Jenny Drnevic, Ruian Ke, Christopher B. Brooke. bioRxiv 858373.
–https://doi.org/10.1101/858373
•Progesterone Receptor Serves the Ovary as a Trigger of Ovulation and a Terminator of Inflammation. Park CJ, Lin PC, Zhou S, et al. Cell Rep. 2020;31(2):107496.
–https://doi.org/10.1016/j.celrep.2020.03.060
PacBio Sequel II is here! (January 2020)
Our new shiny Sequel II is here. We believe that this instrument is a game changer for denovo assembly and annotation of eukaryotic genomes, for sequencing of full-length 16S from metagenomic samples, sequencing of PCR amplicons or DNA fragments with high accuracy and full characterization of transcriptomes, including splice variants . Let us know if you have any project that can benefit from long highly accurate/error-corrected reads.
Single-Cell Genomics and Spatial Genomics Seminar Sponsored by the Sequencing Lab.
Please, attend this seminar on Sept 18th at 612 IGB. We will be covering topics of interest regarding single-cell transcriptomics, single-cell ATAC-seq and Spatial transcriptomics.
See more information in this flyer.
Please, register using this link so that we can plan for the right amount of pizza.
Second NovaSeq 6000 (August 2019)
We just received our second NovaSeq 6000. The addition of this instrument, the latest technology from Illumina, will ensure the fastest possible turnaround time.
Our three HiSeq have been officially retired.


Next time you think watermelon, think Illinois!
https://www.ncbi.nlm.nih.gov/pubmed/31022325
NEW FLOWCELLS FOR THE NOVASEQ (February 2019)
New flowcells have been released for the NovaSeq that effectively make the HiSeq 2500 and HiSeq 4000 obsolete. These flowcells produce a much higher output that the HiSeq systems and the highest possible quality for the lowest cost per Gbase of any sequencing system.
Also, the runs are very short. Our usual turnaround time for the sequencing of libraries constructed by the user is 1 week, sometimes could be up to 2 weeks.
Please, let us know of any questions regarding our NovaSeq library and sequencing services.
Great success in our booth at the 2019 Plant and Animal Genome meeting (January 2019).
We showcased the University of Illinois NGS capabilities.


SINGLE CELL GENOMICS SEMINAR AND GRANT SUPPORT! (October 17th, 2018)
Description:
The DNA Services lab is hosting a lunch seminar on the exciting applications of single cell genomics! We hope you can join us for an informative seminar on single cell RNA sequencing, including new single-cell applications of Immune Profiling and ATAC-Seq.
Please register (see link below) so that we can ensure seating and pizza for all.
See information below regarding a program to support two 2 single-cell projects for UIUC faculty grant proposals.
Featured Speakers:
Andrew Steelman, PhD, Assistant Professor, University of Illinois - 'Differences in glial activation during upper-respiratory viral infection are revealed using 10x single cell RNA-seq'
Dr. Steelman will present an overview of a 10x single-cell experiment performed in collaboration with Dr. Romana Nowak on influenza infected versus non-infected mice glial cells, discussing his techniques for cell preparation and results including identification of cell populations, percentages of cells expressing selective genes and downstream analysis of the dataset.
Melissa Leone, Sales Executive, 10x Genomics
* Reveal cellular heterogeneity and profile unique cell types individually with the Chromium Single Cell Gene Expression Solution
* Simultaneously examine the cellular context of the adaptive immune response and immune repertoires of T and B cells in human or mouse on a cell-by-cell basis with the Chromium Single Cell Immune Profiling Solution
* Perform single cell epigenomics with the Chromium Single Cell Assay for Transposase Accessible Chromatin (ATAC) Solution
Please Register, seating is limited: https://10x_seminar_univofillinois.eventbrite.com
Date/time: October 17th, at 11:45am to 1pm (seminar starts at noon)
Location: 612 IGB (lower floor of gatehouse)
Questions?
Contact Alvaro Hernandez aghernan@illinois.edu or Chris Wright clwright@illinois.edu
10x Genomics and the Roy J. Carver Biotechnology Center are Announcing a Single-Cell Grant Program for Faculty at the UIUC
Description:
10x Genomics, in collaboration with the Roy J. Carver Biotechnology Center, is pleased to announce a pilot award to provide library preparation, sequencing and preliminary bioinformatics analysis for Single Cell 3’ Gene Expression or 5’ Gene Expression and Immune Profiling. This grant will support up to 2 projects consisting of 2 single cell samples per project.
Objective:
The goal of this program is to support UIUC faculty as they seek to generate pilot data in support of future grant applications to the NIH, NSF, USDA and other agencies. Furthermore, it seeks to enable larger research initiatives using 10x Genomics technologies, sequencing and bioinformatic services within the CBC DNA Sequencing and HPCBio facilities.
Eligibility:
- Applicants must be UIUC faculty and must be planning to submit a grant application to support a broader research aim for which they need pilot data.
- Applicants may not hold current awards directly related to the proposed research.
Apply:
To apply, please present a one-page proposal and full contact information by December 14th, 2018. Selected investigators will be notified by December 31st, 2018, and samples must be submitted by February 28th, 2019. Applications must be submitted through this link.
Please send any questions regarding this program to Alvaro G. Hernandez aghernan@illinois.edu or Chris Wright clwright@illinois.edu
Selection of projects will be done by a committee that does not include personnel from the sequencing laboratory.
First NovaSeq 6000 (February 2018)
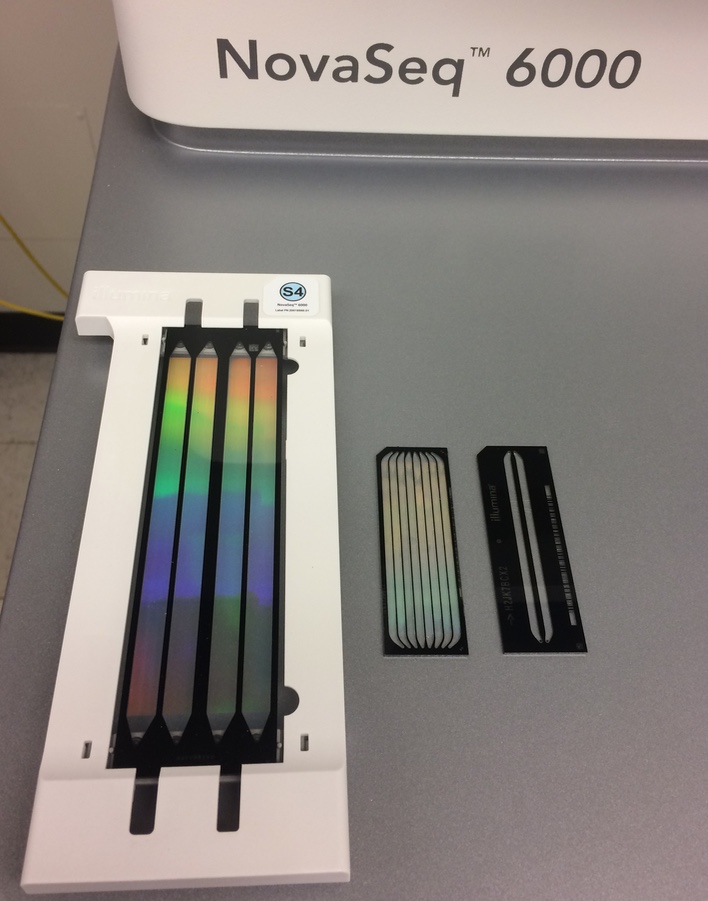
Our NovaSeq 6000 has been installed and it is producing GREAT data. Each lane produces ~ 5 billion reads 150nt in length (2.5 billion clusters sequenced from both ends) with amazing quality scores. This is a cost effective system for projects that require high depth or that have a large number of samples. It has an output equivalent to over 6 lanes in the HiSeq 4000, but for a portion of the cost. The first sequenced projects included several single-cell transcriptomics, 10x Genomics libraries for de novo genome assemblies and shotgun genomics libraries.
This picture shows flowcells from the NovaSeq 6000, HiSeq 4000, and HiSeq 2500, in that order:
Oxford Nanopore GridION (November 2017)
We received the GridION (Oxford Nanopore)! Let us know if you have any project for direct RNA sequencing, de novo assembly of microbial and larger genomes, sequencing of full length 16S or ITS or other projects that can benefit from very long sequence reads (up to 120kb) and single-molecule sequencing.

September 28th, 2017. Illumina seminar.
Illumina RNAseq and Library Construction Seminar Sponsored by the UIUC DNA Sequencing Laboratory
Please join us for an informative seminar. There will be two separate sessions followed by an informal office hour led by
a field applications scientist. Researchers are invited to attend one or all of the sessions.
There is no charge to attend this event, however ***registration is required***
Register now. Space is limited.
***To register please scan the QR Code, or go to:
http://eventregistration.illumina.com/UIUC
9:15 AM - Session I: Introduction to Sequencing Based Methods to Assess Gene Expression and Regulation
This presentation will provide an overview of traditional RNA-Seq applications for surveying the transcribed regions of
the genome; and provide great utility in assessing differential gene expression. We will also discuss how insight into
gene regulation can be achieved through ChIP-Seq and methylation sequencing methods.
10:00 AM - Session II: Best Practices for Illumina Library Preparations
An Illumina Field Applications Scientist will discuss what goes into generating a successful library for sequencing on
all Illumina platforms. The discussion will include quality control of DNA/RNA input, detection and removal of adapter
dimer, the importance of utilizing dual index strategies, and index pooling guidelines. The differences between the
Illumina sequencing platforms will be highlighted to help identify what sequencer is appropriate for specific projects.
11:00 AM - Noon - Session III: Office Hours
Come and meet the UIUC Sequencing and Illumina team and ask questions about sequencing methods.
Speakers Include:
* Alasdair Reid, PhD, Sr. Sales Specialist, Sequencing and Data Analysis, Illumina, Inc.
* Spence Fast, Field Applications Scientist, Illumina, Inc.
Refreshments will be served.*
Date: Thursday, September 28, 2017
Time: 9:00 AM-12:00 PM
Location: University of Illinois at Urbana-Champaign
1206 W. Gregory Drive
612 IGB (Basement of Gatehouse Building)
Urbana, Illinois 61801
For questions, contact Alvaro G. Hernandez at aghernan@illinois.edu<mailto:aghernan@illinois.edu>
April 18th, 2017
Our 10x Genomics Chromium system is up and running! This is a great system for de novo assembly of eukaryotic genomes, genome phasing and characterization of large structural variations. The linked reads offer capabilities that cannot be obtained with short reads alone. It can also do 3' transcriptome libraries from up to 10,000 cells per channel and one chip has 8 channels. Feel free to ask for more information.
February 10th, 2017
We continue to produce complete assemblies of bacterial and viral genomes by sequencing the DNA with Oxford Nanopore and performing error correction with Illumina reads. If you have a bacteria, virus or any small genome that needs a complete assembly let us know.
February 7th, 2017
We are hiring! We have two openings for Research Specialist positions, see add here:
https://jobs.illinois.edu/search-jobs/job-details?jobID=75358&job=resea…
December 19th, 2016
We placed an order for a 10x Genomics Chromium system. This will be used for denovo genome assembly, whole-genome phasing and single-cell transcriptomics. More information here: https://www.10xgenomics.com/
September 6, 2016
Our second HiSeq 4000 has been received. We have consistently been producing between 700 and 800 million paired-reads per lane with the HiSeq 4000. This is over 105 Gbases per lane! We have validated shotgun genomic, RNAseq, bisulfite-converted, RRBS, ChipSeq, small RNA, mate-pair and other libraries in this instrument.
April 25th, 2016
Our first run on the HiSeq 4000 produced 630 to 780 million reads per lane!. With reads being 150nt in length, this is from 94 Gbases to 117 Gbases per lane!, over 30x of a human genome in just one lane. This instrument is great for sequencing of shotgun genomic libraries with fragments ~ 400bp in length and for RNAseq libraries. We will be testing other libraries soon.
April 7th, 2016
Our first HiSeq 4000 is here and getting installed. The HiSeq 4000 is a unit of the HiSeq x10. More updates on this new instrument soon.
April 5th, 2016
We started using Oxford Nanopore for sequencing of archaeal and bacterial genomes with great success! The typical output we get from a MInion MK I flowcell is 15 to 20 thousand reads 1kb to 40kb in length. Please, ask us about this technology.
News reports from sequencing done in our lab:
http://vetmed.illinois.edu/pet_column/new-dog-flu-invites-closer-look-at-viruses-vaccines/
http://wewseq.wix.com/consortium
Day: March, Friday 13th from noon to 1pm. Room 612 IGB.
Title: Next Generation Sequencing at the UIUC DNA Sequencing Laboratory: Applications and Capabilities
Please join us to learn about the very latest next-generation sequencing chemistries, instruments and services at your campus’ DNA sequencing laboratory. We will give an overview of various applications of next-gen sequencing and what resources are available to accomplish your projects, from whole genome sequencing, gene expression profiling, ChipSeq, small RNAs, methylation, metagenomics, microbial community profiling and custom applications. There will be time for questions and answers so come prepared to inquire about the best strategies for your projects. Pizza and refreshments will be provided.
Speakers: Alvaro G. Hernandez, Ph.D. Director of DNA Services
Chris L. Wright, Assistant Director of DNA Services
Seminar sponsored by the DNA Sequencing Laboratory
Date: Tuesday March 17th
Location: ERML room 128
Introducing the MiSeq FGx Forensic Genomics System.
The MiSeq FGx enables forensic DNA testing for criminal casework, forensic profiling for DNA databases, mtDNA analysis to identify missing persons, paternity testing and other forensic applications. This is done in one easy sample prep system that interrogates 200 genetic markers in a single workflow, eliminating the need for multiple STR kits. The MiSeq FGx system includes the ForenSeq Universal Analysis Software, which features a comprehensive suite of library management and analytical tools.
Presenters: Danny Hall and Ann Allison from Illumina.
Watch video from this link:
Download presentations from this link: